
Wellcome to Maximimi's library,
You can find here all papers liked or uploaded by Maximimitogether with brief user bio and description of her/his academic activity.
[Link to my homepage](https://sites.google.com/view/danisch/home)
## I will read the following papers.
- [Quasi-Succinct Indices](https://papers-gamma.link/paper/130)
- [PageRank as a Function of the Damping Factor](https://papers-gamma.link/paper/106)
- [Graph Stream Algorithms: A Survey](https://papers-gamma.link/paper/102)
- [Network Sampling: From Static to Streaming Graphs](https://papers-gamma.link/paper/122)
- [The Protein-Folding Problem, 50 Years On](https://papers-gamma.link/paper/78)
- [Computational inference of gene regulatory networks: Approaches, limitations and opportunitie](https://papers-gamma.link/paper/77)
- [Graph complexity analysis identifies an ETV5 tumor-specific network in human and murine low-grade glioma](https://papers-gamma.link/paper/79)
- [Gene Networks in Plant Biology: Approaches in Reconstruction and Analysis](https://papers-gamma.link/paper/76)
- [The non-convex Burer–Monteiro approach works on smooth semidefinite programs](https://papers-gamma.link/paper/80)
- [Solving SDPs for synchronization and MaxCut problems via the Grothendieck inequality](https://papers-gamma.link/paper/81)
- [Influence maximization in complex networks through optimal percolation](https://papers-gamma.link/paper/70)
- [Motifs in Temporal Networks](https://papers-gamma.link/paper/61)
- [Deep Sparse Rectifier Neural Networks](https://papers-gamma.link/paper/69)
- [Sparse Convolutional Neural Networks](https://papers-gamma.link/paper/67)
- [A fast and simple algorithm for training neural probabilistic language models](https://papers-gamma.link/paper/58)
- [Adding One Neuron Can Eliminate All Bad Local Minima](https://papers-gamma.link/paper/71)
## I read the following papers.
### 2019-2020
- [Fast and High Quality Multilevel Scheme for Partitioning Irregular Graphs](https://papers-gamma.link/paper/162)
- [Karp-Sipser based kernels for bipartite graph matching](https://papers-gamma.link/paper/160)
- [Speedup Graph Processing by Graph Ordering](https://papers-gamma.link/paper/159)
- [Tree Sampling Divergence: An Information-Theoretic Metric for Hierarchical Graph Clustering](https://papers-gamma.link/paper/154)
### 2018-2019:
- [SWeG: Lossless and Lossy Summarization of Web-Scale Graphs](https://papers-gamma.link/paper/139)
- [Smoothed Analysis: An Attempt to Explain the Behavior of Algorithms in Practice](https://papers-gamma.link/paper/129)
- [Are stable instances easy?](https://papers-gamma.link/paper/128)
- [Hierarchical Taxonomy Aware Network Embedding](https://papers-gamma.link/paper/116)
- [Billion-scale Network Embedding with Iterative Random Projection](https://papers-gamma.link/paper/110)
- [HARP: Hierarchical Representation Learning for Networks](https://papers-gamma.link/paper/109/)
- [Layered Label Propagation: A MultiResolution Coordinate-Free Ordering for Compressing Social Networks](https://papers-gamma.link/paper/105)
### 2017-2018:
- [Link Prediction in Graph Streams](https://papers-gamma.link/paper/101)
- [The Community-search Problem and How to Plan a Successful Cocktail Party](https://papers-gamma.link/paper/74)
- [A Nonlinear Programming Algorithm for Solving Semidefinite Programs via Low-rank Factorization](https://papers-gamma.link/paper/55)
- [Deep Learning](https://papers-gamma.link/paper/68)
- [Reducing the Dimensionality of Data with Neural Networks](https://papers-gamma.link/paper/65)
- [Representation Learning on Graphs: Methods and Applications](https://papers-gamma.link/paper/60)
- [Improved Approximation Algorithms for MAX k-CUT and MAX BISECTION](https://papers-gamma.link/paper/56)
- [Cauchy Graph Embedding](https://papers-gamma.link/paper/53)
- [Phase Transitions in Semidefinite Relaxations](https://papers-gamma.link/paper/57)
- [Graph Embedding Techniques, Applications, and Performance: A Survey](https://papers-gamma.link/paper/52)
- [VERSE: Versatile Graph Embeddings from Similarity Measures](https://papers-gamma.link/paper/48)
- [Hierarchical Clustering Beyond the Worst-Case](https://papers-gamma.link/paper/45)
- [Scalable Motif-aware Graph Clustering](https://papers-gamma.link/paper/18)
- [Practical Algorithms for Linear Boolean-width](https://papers-gamma.link/paper/40)
- [New Perspectives and Methods in Link Prediction](https://papers-gamma.link/paper/28/New%20Perspectives%20and%20Methods%20in%20Link%20Prediction)
- [In-Core Computation of Geometric Centralities with HyperBall: A Hundred Billion Nodes and Beyond](https://papers-gamma.link/paper/37)
- [Diversity is All You Need: Learning Skills without a Reward Function](https://papers-gamma.link/paper/36)
- [When Hashes Met Wedges: A Distributed Algorithm for Finding High Similarity Vectors](https://papers-gamma.link/paper/23)
- [Fast Approximation of Centrality](https://papers-gamma.link/paper/35/Fast%20Approximation%20of%20Centrality)
- [Indexing Public-Private Graphs](https://papers-gamma.link/paper/19/Indexing%20Public-Private%20Graphs)
- [On the uniform generation of random graphs with prescribed degree sequences](https://papers-gamma.link/paper/26/On%20the%20uniform%20generation%20of%20random%20graphs%20with%20prescribed%20d%20egree%20sequences)
- [Linear Additive Markov Processes](https://papers-gamma.link/paper/21/Linear%20Additive%20Markov%20Processes)
- [ESCAPE: Efficiently Counting All 5-Vertex Subgraphs](https://papers-gamma.link/paper/17/ESCAPE:%20Efficiently%20Counting%20All%205-Vertex%20Subgraphs)
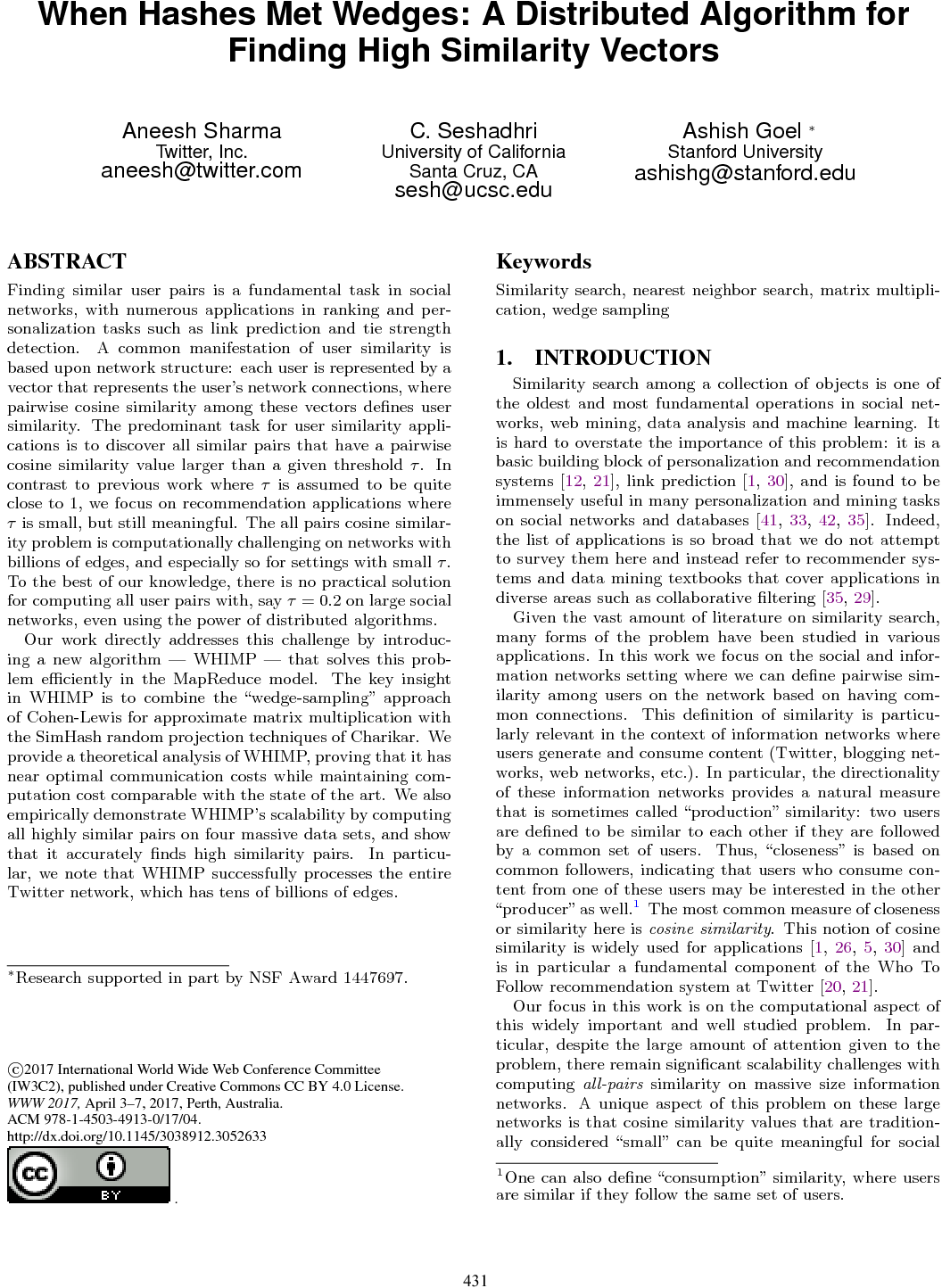
- [The k-peak Decomposition: Mapping the Global Structure of Graphs](https://papers-gamma.link/paper/16/The%20k-peak%20Decomposition:%20Mapping%20the%20Global%20Structure%20of%20Graphs)
- [A Fast and Provable Method for Estimating Clique Counts Using Turán’s Theorem](https://papers-gamma.link/paper/24)
☆
2
Comments:
Just a few remarks from me,
### Definition of mountain
Why is the k-peak in the k-mountain ? Why is the core number of a k-peak changed when removing the k+1 peak ?
###More informations on k-mountains
Showing a k-mountain on figure 2 or on another toy exemple would have been nice.
Besides, more theoretical insights on k-mountains are missing : complexity in time and space of the algorithm used to find the k-mountains ?
How many k-mountains include a given node ?
Less than the degeneracy of the graph but maybe it's possible to say more.
Regarding the complexity of the algorithm used to build the k-mountains, I'm Not sure I understand everything but when the k-peak decomposition is done, for each k-contour, a new k-core decomposition must be run on the subgraph obtained when removing the k-contour. Then it must be compared whith the k-core decomposition of the whole graph. All these operations take time so the complexity of the algorithm used to build the k-mountains is I think higher than the complexity of the k-peak decomposition.
Besides multiple choices are made for building the k-mountains, it seems to me that this tool deserves a broader analysis.
Same thing for the mountains shapes, some insights are missing, for instance mountains may have an obvious staircase shape, does it mean something ?
###6.2 _p_
I don't get really get what is p?
###7.2 is unclear
but it may be related to my ignorance of what is a protein.
### Lemma 2 (bis):
Lemma 2 (bis): Algorithm 1 requires $O(\delta\cdot(N+M))$ time, where $\delta$ is the degeneracy of the input graph.
Proof: $k_1=\delta$ and the $k_i$ values must be unique non-negative integers, there are thus $\delta$ distinct $k_i$ values or less. Thus, Algorithm 1 enters the while loop $\delta$ times or less leading to the stated running time.
We thus have a running time in $O(\min(\sqrt{N},\delta)\cdot (M+N))$.
Note that $\delta \leq \sqrt{M}$ and $\delta<N$ and in practice (in large sparse real-world graphs) it seems that $\delta\lessapprox \sqrt{N}$.
### k-core decomposition definition:
The (full) definition of the k-core decomposition may come a bit late. Having an informal definition of the k-core decomposition (not just a definition of k-core) at the beginning of the introduction may help a reader not familiar with it.
### Scatter plots:
Scatter plots: "k-core value VS k-peak value" for each node in the graph are not shown. This may be interesting.
Note that scatter plots: "k-core value VS degree" are shown in "Kijung, Eliassi-Rad and Faloutsos. CoreScope: Graph Mining Using k-Core Analysis. ICDM2016" leading to interesting insights on graphs. Something similar could be done with k-peak.
###Experiments on large graphs:
As the algorithm is very scalable: nearly linear time in practice (on large sparse real-world graphs) and linear memory. Experiments on larger graphs e.g. 1G edges could be done.
### Implementation:
Even though the algorithm is very easy to implement, a link to a publicly available implementation would make the framework easier to use and easier to improve/extend.
### Link to the densest subgraph:
The $\delta$-core (with $\delta$ the degeneracy of the graph) is a 2-approximation of the densest subgraph (here the density is defined as the average degree divided by 2) and thus the core decomposition can be seen as a (2-)approximation of the density friendly decomposition.
- "Density-friendly graph decomposition". Tatti and Gionis. WWW2015.
- "Large Scale Density-friendly Graph Decomposition via Convex Programming". Danisch et al. WWW2017.
Having this in mind, the k-peak decomposition can be seen as an approximation of the following decomposition:
- 1 Find the densest subgraph.
- 2 Remove it from the graph (along with all edges connected to it).
- 3 Go to 1 till the graph is empty.
### Faster algorithms:
Another appealing feature of the k-core decomposition is that it is used to make faster algorithm. For instance, it is used in https://arxiv.org/abs/1006.5440 and to https://arxiv.org/abs/1103.0318 list all maximal cliques efficiently in sparse real-world graphs.
Can the k-peak decomposition be used in a similar way to make some algorithms faster?
### Section 6.2 not very clear.
### Typos/Minors:
- "one can view the the k-peak decomposition"
- "et. al", "et al"
- "network[23]" (many of these)
- "properties similar to k-core ."
☆
2
Comments:
This paper won the 2018 SEOUL TEST OF TIME AWARD: http://www.iw3c2.org/updates/CP_TheWebConferenceSeoul-ToT-Award-VENG-2018.pdf
### Concerns about "primary pair":
Does a primary pair exist for each n-ary relation? The following example of "NobelPrize" and "AlbertEinstein" given in the paper does not work for "Marie Curie" who won two Nobel prizes according to Wikipedia: https://en.wikipedia.org/wiki/Marie_Curie
" However, this method cannot deal with additional arguments to relations that were designed to be binary. The
YAGO model offers a simple solution to this problem: It is based on the assumption that for each n-ary relation, a
primary pair of its arguments can be identified. For example, for the above won-prize-in-year-relation, the pair of the person and the prize could be considered a primary pair. The primary pair can be represented as a binary fact with a fact identifier: #1 : AlbertEinstein hasWonPrize NobelPrize
All other arguments can be represented as relations that hold between the primary pair and the other argument: #2 : #1 time 1921
"

Comments: